I have a little secret: I don’t like the terminology, notation, and style of writing in statistics. I find it unnecessarily complicated. This shows up when trying to read about Markov Chain Monte Carlo methods. Take, for example, the abstract to the Markov Chain Monte Carlo article in the Encyclopedia of Biostatistics.
Markov chain Monte Carlo (MCMC) is a technique for estimating by simulation the expectation of a statistic in a complex model. Successive random selections form a Markov chain, the stationary distribution of which is the target distribution. It is particularly useful for the evaluation of posterior distributions in complex Bayesian models. In the Metropolis–Hastings algorithm, items are selected from an arbitrary “proposal” distribution and are retained or not according to an acceptance rule. The Gibbs sampler is a special case in which the proposal distributions are conditional distributions of single components of a vector parameter. Various special cases and applications are considered.
I can only vaguely understand what the author is saying here (and really only because I know ahead of time what MCMC is). There are certainly references to more advanced things than what I’m going to cover in this post. But it seems very difficult to find an explanation of Markov Chain Monte Carlo without superfluous jargon. The “bullshit” here is the implicit claim of an author that such jargon is needed. Maybe it is to explain advanced applications (like attempts to do “inference in Bayesian networks”), but it is certainly not needed to define or analyze the basic ideas.
So to counter, here’s my own explanation of Markov Chain Monte Carlo, inspired by the treatment of John Hopcroft and Ravi Kannan.
The Problem is Drawing from a Distribution
Markov Chain Monte Carlo is a technique to solve the problem of sampling from a complicated distribution. Let me explain by the following imaginary scenario. Say I have a magic box which can estimate probabilities of baby names very well. I can give it a string like “Malcolm” and it will tell me the exact probability $ p_{\textup{Malcolm}}$ that you will choose this name for your next child. So there’s a distribution $ D$ over all names, it’s very specific to your preferences, and for the sake of argument say this distribution is fixed and you don’t get to tamper with it.
Now comes the problem: I want to efficiently draw a name from this distribution $ D$. This is the problem that Markov Chain Monte Carlo aims to solve. Why is it a problem? Because I have no idea what process you use to pick a name, so I can’t simulate that process myself. Here’s another method you could try: generate a name $ x$ uniformly at random, ask the machine for $ p_x$, and then flip a biased coin with probability $ p_x$ and use $ x$ if the coin lands heads. The problem with this is that there are exponentially many names! The variable here is the number of bits needed to write down a name $ n = |x|$. So either the probabilities $ p_x$ will be exponentially small and I’ll be flipping for a very long time to get a single name, or else there will only be a few names with nonzero probability and it will take me exponentially many draws to find them. Inefficiency is the death of me.
So this is a serious problem! Let’s restate it formally just to be clear.
Definition (The sampling problem): Let $ D$ be a distribution over a finite set $ X$. You are given black-box access to the probability distribution function $ p(x)$ which outputs the probability of drawing $ x \in X$ according to $ D$. Design an efficient randomized algorithm $ A$ which outputs an element of $ X$ so that the probability of outputting $ x$ is approximately $ p(x)$. More generally, output a sample of elements from $ X$ drawn according to $ p(x)$.
Assume that $ A$ has access to only fair random coins, though this allows one to efficiently simulate flipping a biased coin of any desired probability.
Notice that with such an algorithm we’d be able to do things like estimate the expected value of some random variable $ f : X \to \mathbb{R}$. We could take a large sample $ S \subset X$ via the solution to the sampling problem, and then compute the average value of $ f$ on that sample. This is what a Monte Carlo method does when sampling is easy. In fact, the Markov Chain solution to the sampling problem will allow us to do the sampling and the estimation of $ \mathbb{E}(f)$ in one fell swoop if you want.
But the core problem is really a sampling problem, and “Markov Chain Monte Carlo” would be more accurately called the “Markov Chain Sampling Method.” So let’s see why a Markov Chain could possibly help us.
Random Walks, the “Markov Chain” part of MCMC
Markov Chain is essentially a fancy term for a random walk on a graph.
You give me a directed graph $ G = (V,E)$, and for each edge $ e = (u,v) \in E$ you give me a number $ p_{u,v} \in [0,1]$. In order to make a random walk make sense, the $ p_{u,v}$ need to satisfy the following constraint:
For any vertex $ x \in V$, the set all values $ p_{x,y}$ on outgoing edges $ (x,y)$ must sum to 1, i.e. form a probability distribution.
If this is satisfied then we can take a random walk on $ G$ according to the probabilities as follows: start at some vertex $ x_0$. Then pick an outgoing edge at random according to the probabilities on the outgoing edges, and follow it to $ x_1$. Repeat if possible.
I say “if possible” because an arbitrary graph will not necessarily have any outgoing edges from a given vertex. We’ll need to impose some additional conditions on the graph in order to apply random walks to Markov Chain Monte Carlo, but in any case the idea of randomly walking is well-defined, and we call the whole object $ (V,E, \{ p_e \}_{e \in E})$ a Markov chain.
Here is an example where the vertices in the graph correspond to emotional states.
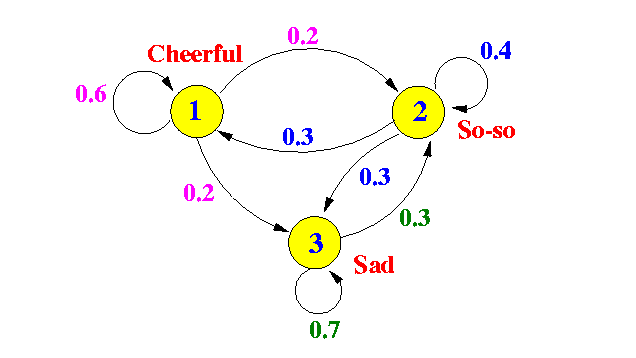
An example Markov chain
In statistics land, they take the “state” interpretation of a random walk very seriously. They call the edge probabilities “state-to-state transitions.” The main theorem we need to do anything useful with Markov chains is the stationary distribution theorem (sometimes called the “Fundamental Theorem of Markov Chains,” and for good reason). What it says intuitively is that for a very long random walk, the probability that you end at some vertex $ v$ is independent of where you started! All of these probabilities taken together is called the stationary distribution of the random walk, and it is uniquely determined by the Markov chain.
However, for the reasons we stated above (“if possible”), the stationary distribution theorem is not true of every Markov chain. The main property we need is that the graph $ G$ is strongly connected. Recall that a directed graph is called connected if, when you ignore direction, there is a path from every vertex to every other vertex. It is called strongly connected if you still get paths everywhere when considering direction. If we additionally require the stupid edge-case-catcher that no edge can have zero probability, then strong connectivity (of one component of a graph) is equivalent to the following property:
For every vertex $ v \in V(G)$, an infinite random walk started at $ v$ will return to $ v$ with probability 1.
In fact it will return infinitely often. This property is called the persistence of the state $ v$ by statisticians. I dislike this term because it appears to describe a property of a vertex, when to me it describes a property of the connected component containing that vertex. In any case, since in Markov Chain Monte Carlo we’ll be picking the graph to walk on (spoiler!) we will ensure the graph is strongly connected by design.
Finally, in order to describe the stationary distribution in a more familiar manner (using linear algebra), we will write the transition probabilities as a matrix $ A$ where entry $ a_{j,i} = p_{(i,j)}$ if there is an edge $ (i,j) \in E$ and zero otherwise. Here the rows and columns correspond to vertices of $ G$, and each column $ i$ forms the probability distribution of going from state $ i$ to some other state in one step of the random walk. Note $ A$ is the transpose of the weighted adjacency matrix of the directed weighted graph $ G$ where the weights are the transition probabilities (the reason I do it this way is because matrix-vector multiplication will have the matrix on the left instead of the right; see below).
This matrix allows me to describe things nicely using the language of linear algebra. In particular if you give me a basis vector $ e_i$ interpreted as “the random walk currently at vertex $ i$,” then $ Ae_i$ gives a vector whose $ j$-th coordinate is the probability that the random walk would be at vertex $ j$ after one more step in the random walk. Likewise, if you give me a probability distribution $ q$ over the vertices, then $ Aq$ gives a probability vector interpreted as follows:
If a random walk is in state $ i$ with probability $ q_i$, then the $ j$-th entry of $ Aq$ is the probability that after one more step in the random walk you get to vertex $ j$.
Interpreted this way, the stationary distribution is a probability distribution $ \pi$ such that $ A \pi = \pi$, in other words $ \pi$ is an eigenvector of $ A$ with eigenvalue 1.
A quick side note for avid readers of this blog: this analysis of a random walk is exactly what we did back in the early days of this blog when we studied the PageRank algorithm for ranking webpages. There we called the matrix $ A$ “a web matrix,” did random walks on it, and found a special eigenvalue whose eigenvector was a “stationary distribution” that we used to rank web pages (this used something called the Perron-Frobenius theorem, which says a random-walk matrix has that special eigenvector). There we described an algorithm to actually find that eigenvector by iteratively multiplying $ A$. The following theorem is essentially a variant of this algorithm but works under weaker conditions; for the web matrix we added additional “fake” edges that give the needed stronger conditions.
Theorem: Let $ G$ be a strongly connected graph with associated edge probabilities $ \{ p_e \}_e \in E$ forming a Markov chain. For a probability vector $ x_0$, define $ x_{t+1} = Ax_t$ for all $ t \geq 1$, and let $ v_t$ be the long-term average $ v_t = \frac1t \sum_{s=1}^t x_s$. Then:
- There is a unique probability vector $ \pi$ with $ A \pi = \pi$.
- For all $ x_0$, the limit $ \lim_{t \to \infty} v_t = \pi$.
Proof. Since $ v_t$ is a probability vector we just want to show that $ |Av_t – v_t| \to 0$ as $ t \to \infty$. Indeed, we can expand this quantity as
$$\displaystyle \begin{aligned} Av_t – v_t &=\frac1t (Ax_0 + Ax_1 + \dots + Ax_{t-1}) – \frac1t (x_0 + \dots + x_{t-1}) \\\ &= \frac1t (x_t – x_0) \end{aligned}$$But $ x_t, x_0$ are unit vectors, so their difference is at most 2, meaning $ |Av_t – v_t| \leq \frac2t \to 0$. Now it’s clear that this does not depend on $ v_0$. For uniqueness we will cop out and appeal to the Perron-Frobenius theorem that says any matrix of this form has a unique such (normalized) eigenvector.
$$\square$$One additional remark is that, in addition to computing the stationary distribution by actually computing this average or using an eigensolver, one can analytically solve for it as the inverse of a particular matrix. Define $ B = A-I_n$, where $ I_n$ is the $ n \times n$ identity matrix. Let $ C$ be $ B$ with a row of ones appended to the bottom and the topmost row removed. Then one can show (quite opaquely) that the last column of $ C^{-1}$ is $ \pi$. We leave this as an exercise to the reader, because I’m pretty sure nobody uses this method in practice.
One final remark is about why we need to take an average over all our $ x_t$ in the theorem above. There is an extra technical condition one can add to strong connectivity, called aperiodicity, which allows one to beef up the theorem so that $ x_t$ itself converges to the stationary distribution. Rigorously, aperiodicity is the property that, regardless of where you start your random walk, after some sufficiently large number of steps $ n$ the random walk has a positive probability of being at every vertex at every subsequent step. As an example of a graph where aperiodicity fails: an undirected cycle on an even number of vertices. In that case there will only be a positive probability of being at certain vertices every other step, and averaging those two long term sequences gives the actual stationary distribution.
Image source: Wikipedia
One way to guarantee that your Markov chain is aperiodic is to ensure there is a positive probability of staying at any vertex. I.e., that your graph has a self-loop. This is what we’ll do in the next section.
Constructing a graph to walk on
Recall that the problem we’re trying to solve is to draw from a distribution over a finite set $ X$ with probability function $ p(x)$. The MCMC method is to construct a Markov chain whose stationary distribution is exactly $ p$, even when you just have black-box access to evaluating $ p$. That is, you (implicitly) pick a graph $ G$ and (implicitly) choose transition probabilities for the edges to make the stationary distribution $ p$. Then you take a long enough random walk on $ G$ and output the $ x$ corresponding to whatever state you land on.
The easy part is coming up with a graph that has the right stationary distribution (in fact, “most” graphs will work). The hard part is to come up with a graph where you can prove that the convergence of a random walk to the stationary distribution is fast in comparison to the size of $ X$. Such a proof is beyond the scope of this post, but the “right” choice of a graph is not hard to understand.
The one we’ll pick for this post is called the Metropolis-Hastings algorithm. The input is your black-box access to $ p(x)$, and the output is a set of rules that implicitly define a random walk on a graph whose vertex set is $ X$.
It works as follows: you pick some way to put $ X$ on a lattice, so that each state corresponds to some vector in $ \{ 0,1, \dots, n\}^d$. Then you add (two-way directed) edges to all neighboring lattice points. For $ n=5, d=2$ it would look like this:
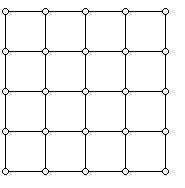
Image credit http://www.ams.org/samplings/feature-column/fcarc-taxi
And for $ d=3, n \in \{2,3\}$ it would look like this:
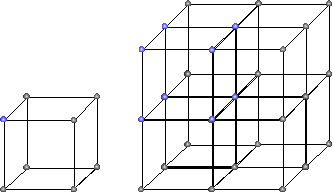
lattice
You have to be careful here to ensure the vertices you choose for $ X$ are not disconnected, but in many applications $ X$ is naturally already a lattice.
Now we have to describe the transition probabilities. Let $ r$ be the maximum degree of a vertex in this lattice ($ r=2d$). Suppose we’re at vertex $ i$ and we want to know where to go next. We do the following:
- Pick neighbor $ j$ with probability $ 1/r$ (there is some chance to stay at $ i$).
- If you picked neighbor $ j$ and $ p(j) \geq p(i)$ then deterministically go to $ j$.
- Otherwise, $ p(j) < p(i)$, and you go to $ j$ with probability $ p(j) / p(i)$.
We can state the probability weight $ p_{i,j}$ on edge $ (i,j)$ more compactly as
$$\displaystyle p_{i,j} = \frac1r \min(1, p(j) / p(i)) \\\ p_{i,i} = 1 – \sum_{(i,j) \in E(G); j \neq i} p_{i,j}$$It is easy to check that this is indeed a probability distribution for each vertex $ i$. So we just have to show that $ p(x)$ is the stationary distribution for this random walk.
Here’s a fact to do that: if a probability distribution $ v$ with entries $ v(x)$ for each $ x \in X$ has the property that $ v(x)p_{x,y} = v(y)p_{y,x}$ for all $ x,y \in X$, the $ v$ is the stationary distribution. To prove it, fix $ x$ and take the sum of both sides of that equation over all $ y$. The result is exactly the equation $ v(x) = \sum_{y} v(y)p_{y,x}$, which is the same as $ v = Av$. Since the stationary distribution is the unique vector satisfying this equation, $ v$ has to be it.
Doing this with out chosen $ p(i)$ is easy, since $ p(i)p_{i,j}$ and $ p(i)p_{j,i}$ are both equal to $ \frac1r \min(p(i), p(j))$ by applying a tiny bit of algebra to the definition. So we’re done! One can just randomly walk according to these probabilities and get a sample.
Last words
The last thing I want to say about MCMC is to show that you can estimate the expected value of a function $ \mathbb{E}(f)$ simultaneously while random-walking through your Metropolis-Hastings graph (or any graph whose stationary distribution is $ p(x)$). By definition the expected value of $ f$ is $ \sum_x f(x) p(x)$.
Now what we can do is compute the average value of $ f(x)$ just among those states we’ve visited during our random walk. With a little bit of extra work you can show that this quantity will converge to the true expected value of $ f$ at about the same time that the random walk converges to the stationary distribution. (Here the “about” means we’re off by a constant factor depending on $ f$). In order to prove this you need some extra tools I’m too lazy to write about in this post, but the point is that it works.
The reason I did not start by describing MCMC in terms of estimating the expected value of a function is because the core problem is a sampling problem. Moreover, there are many applications of MCMC that need nothing more than a sample. For example, MCMC can be used to estimate the volume of an arbitrary (maybe high dimensional) convex set. See these lecture notes of Alistair Sinclair for more.
If demand is popular enough, I could implement the Metropolis-Hastings algorithm in code (it wouldn’t be industry-strength, but perhaps illuminating? I’m not so sure…).
Until next time!
Want to respond? Send me an email, post a webmention, or find me elsewhere on the internet.